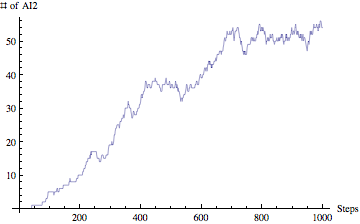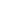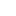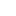I am pleased to post (for the first time) screen shots of our Membrane Computing Agent Base Model. In this post i am focusing on interface mostly. Beside the fact that we want to have an accurate model, we also want a model that is easy to use. So we thought about design issues with our interface. How can we (computer scientists) hide as much code as possible so when a biologist uses our software, they don't freak out.
So lets take a look at our main screen:
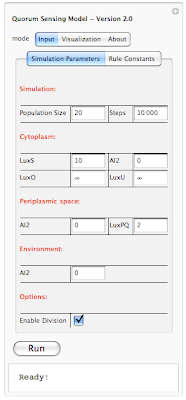
If the size of the picture is small and you have a hard time making out the details, you can always click on the photo and see a full size version. Unfortunately this is the largest size that the blog allows for posting images online. Anyways going back to our interface, on the top you see "Quorum Sensing Model - Version 2.0". So this is our QS model and the version 2.0 is our latest release. We initially started with version 1.0 and gone through 1.1, 1.2, 1.3, ... , 1.7. With versions 1.x we were adding basic functionality to the system. Simply setting up a framework, introducing rules, figuring out how to define rules and so on. In version 2.0 we have build the interface on top of our framework so users can interact with the system without the need of knowing how the code works. You don't need to compile anything, or type a single line of code. This is the beautify of Mathematica. I don't want to get into Mathematica and our implementation; however I want to mentioned that all you need to know to run our program is a shift+enter and that will do the trick. We have also worked on improving run-time speed of our simulation and I am happy to say that version 2.0 is about 5x faster than version 1.0!
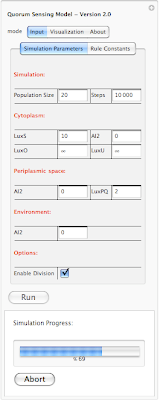
In this example the simulation is 69% completed. Next is looking at rule constants. Each rule in our simulation has a constant associated with it. This constant is used for calculating probability of a reaction. The system is loaded with default values for each constant. However one can modify each value before running the simualtion. Here is an screen shot of rule constants:
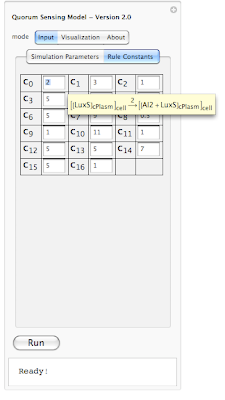 Because it is hard to remember which constant belongs to which rule, we have enabled a tool-tip option so when a user moves the mouse pointer over each constant, its corresponding interaction rule is displayed (as shown above). This should help out the user in associating each constant value with an interaction rule in the system.
Because it is hard to remember which constant belongs to which rule, we have enabled a tool-tip option so when a user moves the mouse pointer over each constant, its corresponding interaction rule is displayed (as shown above). This should help out the user in associating each constant value with an interaction rule in the system. 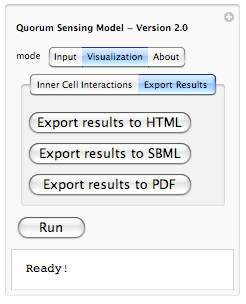 Yes, we are planning to integrate the ability of exporting results from the simulation to HTML (for posting online), SBML (Systems Biology Markup Language - for visualizing metabolic pathways and importing to popular biological modeling tools such as Cell Designer) and PDF (for sharing as well as printing). This is part of our future work for expanding the model and making it even more useful.
Yes, we are planning to integrate the ability of exporting results from the simulation to HTML (for posting online), SBML (Systems Biology Markup Language - for visualizing metabolic pathways and importing to popular biological modeling tools such as Cell Designer) and PDF (for sharing as well as printing). This is part of our future work for expanding the model and making it even more useful.