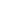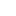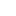The closer the graph gets to 0, the lower the influence this parameter has on the GFP output, and vice versa. According to this graph, the kForward (reaction rates)values 1, 3, and 4 seems the most influential parameters relative to 2 and 5; thus these parameters will be looked at further.
From these results, Vicki performed parameter optimization on them. In parameter optimization function, she was able to play with the individual parameter values in order to come up with a best fit line model to the predicted levels of GFP output. The following is the graphical result of the above function.

Here, the circles represent the made up predicted data, and the colored lines represent each optimized parameters. The optimized rate constants for kForward1, 3, and 4 fit nicely to the pattern of the predicted data; however, the optimized rate constants for kForward 2 and 5 do not, meaning that these parameters do not play a big role in determining the GFP output values.
From these two functions, Vicki would be able to, once we get some lab results, optimize our significant rate constants to fit the behavior of our AI-2 system.
Along with Vicki, Chinee was also working on the parameter sensitivity; however, with a different tool. In Simbiology, Chinee was able to outline the key difference between the initial amount and the reaction rates of the parameter. The following is the graph when all the parameters have a reaction rate value of 1:

The following is the graph when all the parameters have a reaction rate value of 10:

The only difference between the above two graphs are that the lines in the second graph are much more steep than the lines in the first graph. This means that the reaction rate only influences how immediate the reactions happen.
And finally, the graph when all the parameters have an initial amount of 10:

The above graph is very different from the ones above. From this, one can see how initial amounts of parameter play a much bigger role than the rate constants of each parameters.
I, Kevin, and Carol attempted to fix some biological misunderstandings within the reactions, and produce a graph that displays the pattern of each parameters. The following is the graph:

The patterns observed are reasonable and match what was expected. The lab data is needed to get an accurate model of our system; however, from it we can still test the sensitivity of each parameters by changing them one by one and simulating the results.
We hope to get some lab data by this week.